利用cafe5进行基因家族扩张收缩分析 测试于2023年9月1日
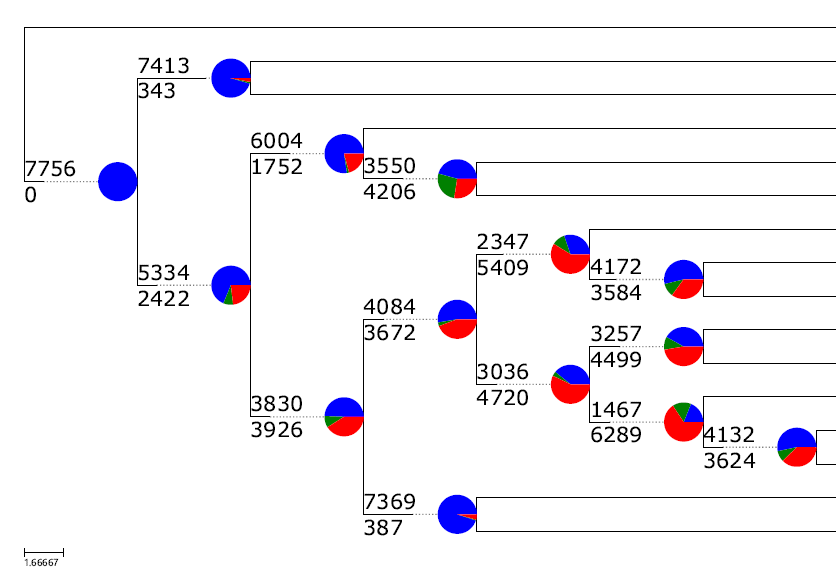
基于orthofinder直系同源聚类的结果,可以看在树的特定节点上哪些基因家族发生了扩张与收缩
软件安装
1 2 3 4 5 6 conda activate biosoft conda install -c bioconda cafe cafe5 -h
准备文件包括1、带有分歧时间的树,由MCMCtree产生;2、直系同源基因家族的聚类情况,由orthofinder结果产生;
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 grep "UTREE 1 =" FigTree.tre | sed -E -e "s/\[[^]]*\]//g" -e "s/[ \t]//g" -e "/^$/d" -e "s/UTREE1=//" > tree.txt awk -v OFS="\t" '{$NF=null;print $1,$0}' Orthogroups.GeneCount.tsv |sed -E -e 's/Orthogroup/desc/' -e 's/_[^\t]+//g' > orthomcl2cafe.tab perl orthoMCL2cafe.pl Orthogroups.txt > orthomcl2cafe.tab python ~/scripts/cafetutorial_clade_and_size_filter.py -i orthomcl2cafe.tab -o gene_family_filter.txt -s
运行cafe5软件
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 cafe5 -i orthomcl2cafe.tab -t tree.txt -p -o singlelambda Families with largest size differentials: OG0000385: 97 OG0000296: 97 OG0000029: 96 OG0000030: 95 OG0000016: 93 OG0000142: 92 OG0000232: 91 OG0000035: 91 OG0000155: 89 OG0000115: 89 OG0000027: 88 OG0000079: 86 OG0000034: 83 OG0000044: 82 OG0000055: 80 OG0000067: 79 OG0000943: 78 OG0000050: 77 OG0000110: 76 OG0000088: 76 You may want to try removing the top few families with the largest difference between the max and min counts and then re-run the analysis. Failed to initialize any reasonable values vi families_largest.txt sed -i 's/:.*//g' families_largest.txt for i in `cat families_largest.txt`;do sed -i "/$i /d" orthomcl2cafe.tab;done (grep -v -f family.remove.id.txt input.tab >input.tab.new) cafe5 -i orthomcl2cafe.tab -t tree.txt -p -o singlelambda cafe5 -i orthomcl2cafe.tab -t tree.txt -p -k 2 -l 0.0001 -o k2p cafe5 -i orthomcl2cafe.tab -t tree.txt -p -k 3 -l 0.0001 -o k3p cafe5 -i orthomcl2cafe.tab -t tree.txt -p -k 4 -l 0.0001 -o k4p cafe5 -i orthomcl2cafe.tab -t tree.txt -p -k 5 -l 0.0001 -o k5p
在结果中的Gamma_results.txt文件里查看lnL值,选择该值最大的k值运行结果作为后续分析的文件
接下来提取感兴趣节点的基因家族序列
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 cat ../Gamma_change.tab |cut -f1,37|grep "+[1-9]" > cr.expanded grep "<35>\*" ../Gamma_asr.tre > cr_significant_trees.tre grep -E -o "OG[0-9]+" cr_significant_trees.tre > cr_significant.ogs awk '$2 <0.05 {print $1}' ../Gamma_family_results.txt >p0.05_significant.ogs grep -f cr_significant.ogs p0.05_significant.ogs > cr_p0.05_significant.ogs grep -f cr_p0.05_significant.ogs cr.expanded |cut -f1 > cr.expanded.significant grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep "amil" |sort -k 1.3n |uniq > cr.amil.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep adig |sort -k 1.3n |uniq > cr.adig.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep aequ |sort -k 1.3n |uniq > cr.aequ.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep afen |sort -k 1.3n |uniq > cr.afen.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep alor |sort -k 1.3n |uniq > cr.alor.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep amil |sort -k 1.3n |uniq > cr.amil.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep apall |sort -k 1.3n |uniq > cr.apall.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep atene |sort -k 1.3n |uniq > cr.atene.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep atenu |sort -k 1.3n |uniq > cr.atenu.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep dgig |sort -k 1.3n |uniq > cr.dgig.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep disc |sort -k 1.3n |uniq > cr.disc.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep edia |sort -k 1.3n |uniq > cr.edia.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep fung |sort -k 1.3n |uniq > cr.fung.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep gasp |sort -k 1.3n |uniq > cr.gasp.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep gfas |sort -k 1.3n |uniq > cr.gfas.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep hvul |sort -k 1.3n |uniq > cr.hvul.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep nvec |sort -k 1.3n |uniq > cr.nvec.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep ofav |sort -k 1.3n |uniq > cr.ofav.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep pdae |sort -k 1.3n |uniq > cr.pdae.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep pdam |sort -k 1.3n |uniq > cr.pdam.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep plut |sort -k 1.3n |uniq > cr.plut.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep pspe |sort -k 1.3n |uniq > cr.pspe.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep pverr |sort -k 1.3n |uniq > cr.pverr.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep rmue |sort -k 1.3n |uniq > cr.rmue.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep spis |sort -k 1.3n |uniq > cr.spis.expanded.significant.genes grep -f cr.expanded.significant ~/2023-redo-cafe/OrthoFinder/Results_Aug27/Orthogroups/Orthogroups.txt|sed "s/ /\n/g" |grep xeni |sort -k 1.3n |uniq > cr.xeni.expanded.significant.genes seqkit grep -f ../cr.adig.expanded.significant.genes /home/tianzhen/2023-redo-cafe/adig.fasta > cr.adig.expanded.significant.pep.fas seqkit grep -f ../cr.aequ.expanded.significant.genes /home/tianzhen/2023-redo-cafe/aequ.fasta > cr.aequ.expanded.significant.pep.fas seqkit grep -f ../cr.afen.expanded.significant.genes /home/tianzhen/2023-redo-cafe/afen.fasta > cr.afen.expanded.significant.pep.fas seqkit grep -f ../cr.alor.expanded.significant.genes /home/tianzhen/2023-redo-cafe/alor.fasta > cr.alor.expanded.significant.pep.fas seqkit grep -f ../cr.amil.expanded.significant.genes /home/tianzhen/2023-redo-cafe/amil.fasta > cr.amil.expanded.significant.pep.fas seqkit grep -f ../cr.apall.expanded.significant.genes /home/tianzhen/2023-redo-cafe/apall.fasta > cr.apall.expanded.significant.pep.fas seqkit grep -f ../cr.atene.expanded.significant.genes /home/tianzhen/2023-redo-cafe/atene.fasta > cr.atene.expanded.significant.pep.fas seqkit grep -f ../cr.atenu.expanded.significant.genes /home/tianzhen/2023-redo-cafe/atenu.fasta > cr.atenu.expanded.significant.pep.fas seqkit grep -f ../cr.dgig.expanded.significant.genes /home/tianzhen/2023-redo-cafe/dgig.fasta > cr.dgig.expanded.significant.pep.fas seqkit grep -f ../cr.disc.expanded.significant.genes /home/tianzhen/2023-redo-cafe/disc.fasta > cr.disc.expanded.significant.pep.fas seqkit grep -f ../cr.edia.expanded.significant.genes /home/tianzhen/2023-redo-cafe/edia.fasta > cr.edia.expanded.significant.pep.fas seqkit grep -f ../cr.fung.expanded.significant.genes /home/tianzhen/2023-redo-cafe/fung.fasta > cr.fung.expanded.significant.pep.fas seqkit grep -f ../cr.gasp.expanded.significant.genes /home/tianzhen/2023-redo-cafe/gasp.fasta > cr.gasp.expanded.significant.pep.fas seqkit grep -f ../cr.gfas.expanded.significant.genes /home/tianzhen/2023-redo-cafe/gfas.fasta > cr.gfas.expanded.significant.pep.fas seqkit grep -f ../cr.hvul.expanded.significant.genes /home/tianzhen/2023-redo-cafe/hvul.fasta > cr.hvul.expanded.significant.pep.fas seqkit grep -f ../cr.nvec.expanded.significant.genes /home/tianzhen/2023-redo-cafe/nvec.fasta > cr.nvec.expanded.significant.pep.fas seqkit grep -f ../cr.ofav.expanded.significant.genes /home/tianzhen/2023-redo-cafe/ofav.fasta > cr.ofav.expanded.significant.pep.fas seqkit grep -f ../cr.pdae.expanded.significant.genes /home/tianzhen/2023-redo-cafe/pdae.fasta > cr.pdae.expanded.significant.pep.fas seqkit grep -f ../cr.pdam.expanded.significant.genes /home/tianzhen/2023-redo-cafe/pdam.fasta > cr.pdam.expanded.significant.pep.fas seqkit grep -f ../cr.plut.expanded.significant.genes /home/tianzhen/2023-redo-cafe/plut.fasta > cr.plut.expanded.significant.pep.fas seqkit grep -f ../cr.pspe.expanded.significant.genes /home/tianzhen/2023-redo-cafe/pspe.fasta > cr.pspe.expanded.significant.pep.fas seqkit grep -f ../cr.pverr.expanded.significant.genes /home/tianzhen/2023-redo-cafe/pverr.fasta > cr.pverr.expanded.significant.pep.fas seqkit grep -f ../cr.rmue.expanded.significant.genes /home/tianzhen/2023-redo-cafe/rmue.fasta > cr.rmue.expanded.significant.pep.fas seqkit grep -f ../cr.spis.expanded.significant.genes /home/tianzhen/2023-redo-cafe/spis.fasta > cr.spis.expanded.significant.pep.fas seqkit grep -f ../cr.xeni.expanded.significant.genes /home/tianzhen/2023-redo-cafe/xeni.fasta > cr.xeni.expanded.significant.pep.fas cat *.fas > all_sequences_for_anno.fas
参考
hahnlab/CAFE5: Version 5 of the CAFE phylogenetics software (github.com)
分析基因家族扩张和收缩 —— CAFE5 | 生信技工 (yanzhongsino.github.io)